Wound Healing Assay
How to measure wound closure over time using image analysis?
Discover each assay
Click on
How to measure wound closure over time using image analysis?
How to measure migration from circular geometry using image analysis?
WOUND HEALING ASSAY
How to measure wound closure over time using image analysis?
4Dcell technology
SmartPattern Technology
Read-outs
Wound healing, % wound closure, wound closing rate, cell migration, area occupied by cells, image object detection
Standard culture limitation
The wound healing assay is traditionally performed by scratching a cell monolayer to form a cell-free region (wound) and monitor cell migration into the wound. Although easy to set-up and inexpensive, this method creates inconsistent wounds with complicated geometries, which make the images difficult to analyze.
Wound healing assay benefits
The method calculates the area occupied by the cells on the image. As cells migrate over time, the area occupied by the cells increases and the wound region becomes smaller until it is fully covered by cells. The method uses CellProfiler, a free, python-based open source software for biological images analysis. Note that it is also possible to use other softwares or to write a program from scratch to perform the analysis.
Example
In this example, a wound healing assay was set-up on dynamic micropatterns. Mesenchymal stem cells were cultured on a 4DCELL 18 mm dynamic micropatterned glass coverslips, not coated with any ECM protein.
The micropatterned coverslips consisted of a rectangular shape of width 500 um coated with the anti-adhesive agent. 60’000 cells were seeded on the coverslip.
After 24 hours of incubation, the cells were homogeneously patterned in the regions surrounding the wound.
To activate migration into the wound, a high dose (50 uM) of the triggering reagent was added to render the wound region adhesive.

Setting up the wound healing assay using a 4DCELL dynamic micropatterned coverslip.
Data analysis
The rate of cell migration was quantified by measuring the area occupied by cells over time.
First, the background was subtracted from the image using the imageJ built-in ‘’rolling ball’’ algorithm. This allowed to enhance the contrast between cells and background and to correct for uneven illumination.
The image was converted to a grayscale image using the ‘ColorToGray’ CellProfiler module. The ‘Smooth’ module was then applied to the grayscale image to remove artifacts. Next, the ‘IdentifyPrimaryObjects’ and ‘MeasureImageAreaOccupied’ modules were successively applied. The first classifies features on the image into two two classes: object and background, where the cells correspond to objects. The latter measures the area taken by the previously calculated object on the image.

Sequence of modules used in the CellProfiler pipeline for measuring the area occupied by cells on the image. The area occupied by the cells on the original image is 50%.
Results
Images of the substrate were acquired at three timepoints: immediately after triggering cell migration with the reagent, 8 hours after and 24 hours after. A non-treated negative control is also provided. The area occupied by cells was calculated using CellProfiler. On the treated sample, migration into the wound steadily increases and the wound is almost fully closed after 24 hours (98.6% closure). On the control sample the rate of wound closure is less. Partial wound closure on the non-treated sample after 24 hours is likely due to cell proliferation because the regions surrounding the wound are too confluent.

Setting up the wound healing assay using a 4DCELL dynamic micropatterned coverslip.
This simple example illustrates how it is possible to generate and analyze data using the 4DCELL dynamic micropatterned plate. Object detection by image segmentation is a powerful method to quantify the rate of wound closure on the 4DCELL wound healing assay.
DISK MIGRATION ASSAY
How to measure migration from circular geometry using image analysis?
4Dcell technology
SmartPattern Technology
Read-outs
Controlled cell migration, collective cell migration, area occupied by cells, image object detection, circular migration, cell polarity
Standard culture limitation
Migration of a cell population is usually measured with the wound healing assay. However, it is only a large scale assay with a high number of cells in the population, making it difficult to analyse the behaviour of single cells and to distinguish cell migration from cell proliferation.
Wound healing assay benefits
The substrate consists of an array of adhesive circular micropatterns of 500 um. The cells are confined to these circles and fill them as they proliferate. Once they are confluent in the circles, migration is triggered by rendering the outside of the circles newly adhesive. The circles of cells on the substrate can either be studied as single entities or as a whole, a single micropatterned substrate can thus be seen as a multiplexed migration assay containing a certain amount of smaller-scale cell populations. Depending on the radius of a circle, the number of individual circular populations of cells on the subs1trate will vary.
In the present application note, we describe how migration of a small population of cells, consisting of around 200 cells patterned inside a circle of 500 um diameter can be monitored in time using the 4DCELL dynamic micropatterns.
Example
In this example, a circle migration assay was set-up on dynamic micropatterns. Mesenchymal stem cells were cultured on a 4DCELL 18 mm dynamic micropatterned glass coverslips, not coated with any ECM protein. The micropatterned coverslips consisted of an array of circular shapes of diameter 500 um coated with the anti-adhesive agent. 60’000 cells were seeded on the coverslip. After 24 hours of incubation, the cells were homogeneously patterned in the circles and absent in regions separating the circles. To release the cells from their patterns, the region surrounding the circles is rendered adhesive by adding a high dose (50 uM) of the triggering reagent.

Setting up the circle migration assay using a 4DCELL dynamic micropatterned coverslip
Data analysis
Cell migration from circular patterns can be quantified by measuring the area occupied by the spreading cells at different timepoints.
First, the background is subtracted from the image using the imageJ built-in ‘’rolling ball’’ algorithm. This allows to enhance the contrast between cells and background and to correct for uneven illumination.
Then, the image is converted to a grayscale image using the ‘ColorToGray’ CellProfiler module. The ‘Smooth’ module is then applied to the grayscale image to remove artifacts.
Next, the ‘IdentifyPrimaryObjects and ‘MeasureImageAreaOccupied’ modules are successively applied.
The first classifies features on the image into two classes: object and background, where the cells correspond to objects. The latter measures the area taken by the previously calculated object on the image.
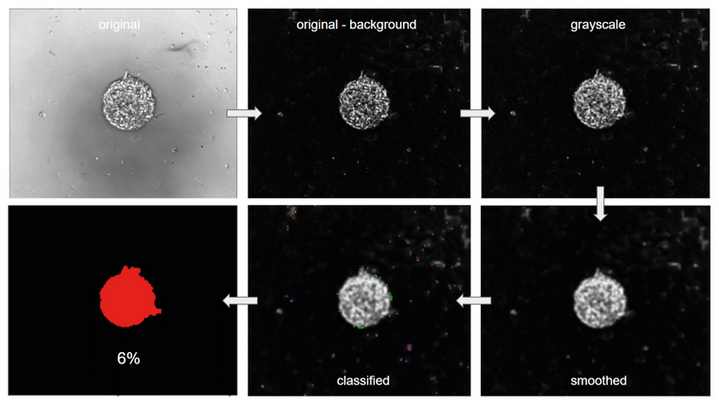
Sequence of modules used in the CellProfiler pipeline for measuring the area occupied by cells on the image
In this example, the area occupied by the cells on the original image is 6%
Results
Images of the substrate were acquired at three timepoints: immediately after triggering cell migration with the reagent, 4 hours, 8 hours and 24 hours after. A non-treated negative control is also provided.
The area occupied by cells was calculated using CellProfiler

Setting up the wound healing assay using a 4DCELL dynamic micropatterned coverslip.
This simple example illustrates how to the 4DCELL dynamic micropatterns can be used to monitor the migration of a small cell population from an initial circular pattern. On the treated sample, migration outside of the 500 um circle steadily increases.
After 24 hours, the cells have migrated to cover 43% of the frame.
At 4 and 8 hours, the spreading of the circle is mostly caused by migration, a process that has a shorter time-scale compared to cell proliferation.
After some time, the combination of both proliferation and migration amplify spreading of the cells onto the substrate.
Interestingly, the cells retain a very regular circular shape as they migrate.
