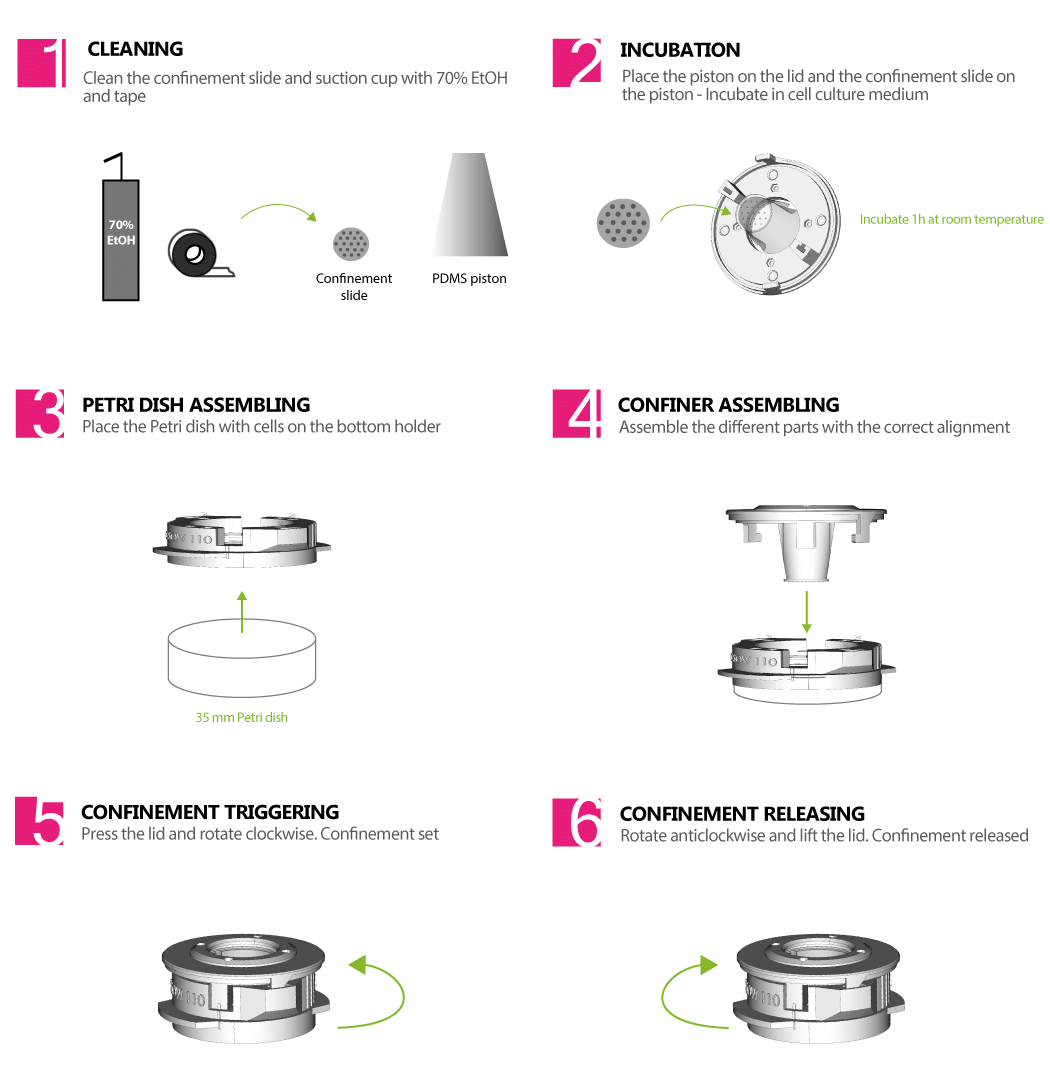
EXPLORE CELL MECHANICS 1-WELL STATIC CONFINER – CSOW 110
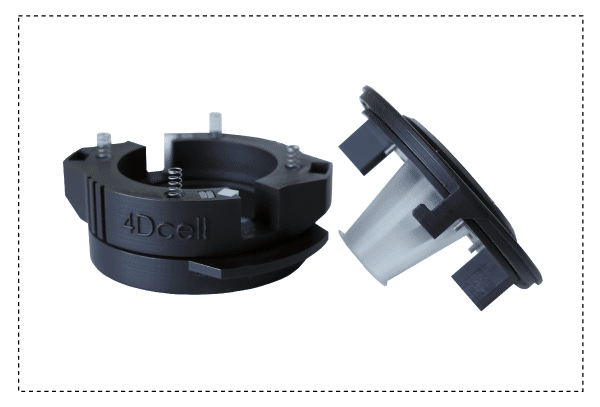
The 1-well static confiner is a portable device that confines cells between two surfaces to a defined micrometer height with nanometer precision. The space between the two surfaces is controlled by using micro PDMS pillars. The micropillars are fabricated on a glass slide, which is attached to a PDMS piston.
The CSOW 110 is the device that controls the piston
> USER FRIENDLY AND CONVENIENT
It is easy to assemble and is portable
> ENABLES HIGH RESOLUTION IMAGING
It fits with glass bottom petri dishes (35 mm) and the observation area is made of optically transparent materials
> ADAPTABLE WITH YOUR MICROSCOPE
It was designed to fit in your microscope
FEATURES & BENEFITS
THE STATIC CELL CONFINER CSOW 110 IS COMPOSED OF:
> 1 x CSOW 110
> 12 x Confinement Slides – 16 mm (diameter) glass slides/cover slips with micro-structures in PDMS (pillars) that enable the confinement
Available confinement heights – from 1, 2, 3, … up to 20 µm (you can choose up to 3 heights to integrate your kit)
> 2 x Confinement Piston in PDMS
IF YOU WANT TO MAKE YOUR SLIDES ADHESIVE TO YOUR CELLS OR NOT (OPTIONAL):
Aliquots of extracellular matrix protein for cell adhesion (for example, fibronectin) in the right buffer solution
Aliquots of anti-adhesive molecule (poly-ethylene glycol) ready to bind on the slides
If this device does not meet your needs, get in touch with us! We can personalize it for you.
VIDEO PROTOCOL TO SET UP PERFECTLY YOUR CSOW 110
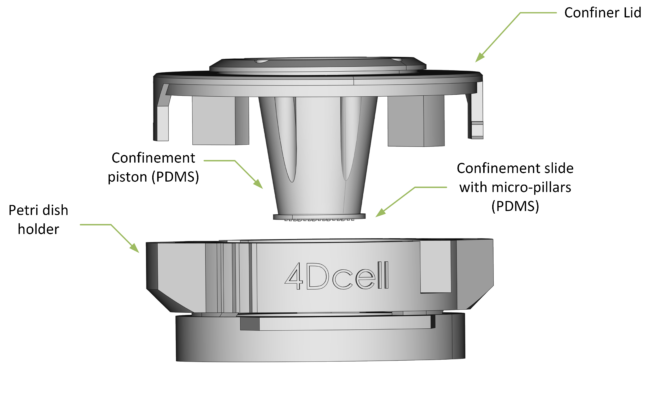
ANATOMY OF THE CSOW 110
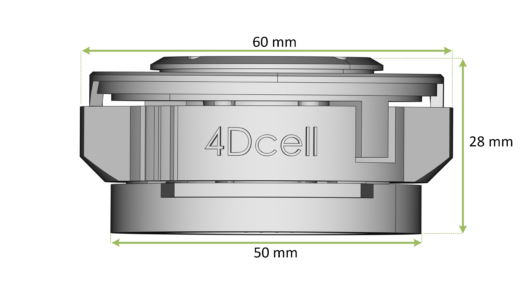
Overall dimensions of the CSOW 110

The lid enables access to the petri dish, allowing cell media to be exchanged while cells are confined.
CANCER
> Migration of metastatic cells
> Cell contractility in mestastasis
> DNA DSB repair (mechanically induced)
> Genomic instability (cell division)
> Separated co-culture
IMMUNOLOGY
> Migration of immune cells
> Imaging of non-adhesive cells
ORGAN PHYSIOLOGY
> Migration of cancer cells
> Cell differenciation with stiffness control
> Wound healing
> Separeted co-culture
> Cell compression response
RARE DISEASES
> Cell nucleus integrity
AGING
> Cell nucleus integrity
> Autophagy related diseases
OBSERVATION OPTIMIZATION
> Imaging of non-adhesive cells
> Planar imaging of organelles
FUNDAMENTAL RESEARCH
> Cell volume (cell cycle)
> Cell stretching response
Video of HeLa cells under confinement using a 4Dcell confiner, going from initial state to extremely confined.
Video of cells dividing under confinement using a 4Dcell confiner
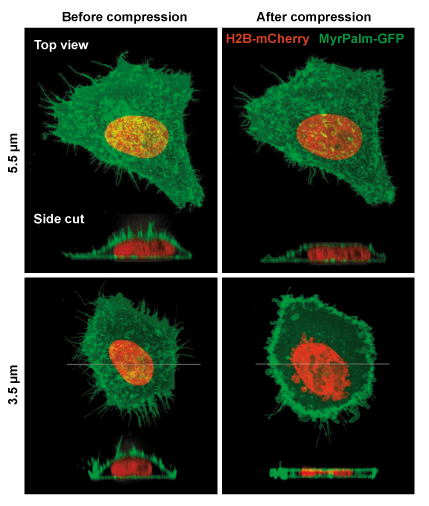
Example of mammalian cells with before and after confinement images with fluorescent proteins
> Confinement and Low Adhesion Induce Fast Amoeboid Migration of Slow Mesenchymal Cells
Y.-J. Liu, M. Piel, Cell, et al., 2015 160(4), 659-672
> Actin flows induce a universal coupling between cell speed and cell persistence
P. Maiuri, R. Voituriez, et al., Cell, 2015 161(2), 374–386
> Geometric friction directs cell migration
M. Le Berre, M. Piel, et al., Physical Review Letter 2013 111, 198101
> Mitotic rounding alters cell geometry to ensure efficient spindle assembly
O. M. Lancaster, B. Baum, et al., Developmental Cell, 2013 25(3), 270-283
> Fine Control of Nuclear Confinement Identifies a Threshold Deformation leading to Lamina Rupture and Induction of Specific Genes
M. Le Berre, J. Aubertin, M. Piel, Integrative Biology, 2012 4 (11), 1406-1414
> Exploring the Function of Cell Shape and Size during Mitosis
C. Cadart, H. K. Matthews, et al., Developmental Cell, 2014 29(2), 159-169
> Methods for Two-Dimensional Cell Confinement
M. Le Berre, M. Piel, et al., 2014, Micropatterning in Cell Biology Part C, Methods in cell biology, 121, 213-29